from simpeg import (
maps, utils, data, optimization, maps, regularization,
inverse_problem, directives, inversion, data_misfit
)
import discretize
from discretize.utils import mkvc, refine_tree_xyz
from simpeg.electromagnetics import natural_source as nsem
import numpy as np
from scipy.spatial import cKDTree
import matplotlib.pyplot as plt
from pymatsolver import Pardiso as Solver
from matplotlib.colors import LogNorm
from ipywidgets import interact, widgets
import warnings
from mtpy import MTCollection
warnings.filterwarnings("ignore", category=FutureWarning)Get Data from MTpy¶
%%time
with MTCollection() as mc:
mc.open_collection("../../data/transfer_functions/yellowstone_mt_collection.h5")
out = mc.apply_bbox(*[-111.4, -109.85, 44, 45.2])
mc.mth5_collection.tf_summary.summarize()
mt_data=mc.to_mt_data(bounding_box=[-111.4, -109.85, 44, 45.2])24:10:18T09:59:43 | INFO | line:777 |mth5.mth5 | close_mth5 | Flushing and closing ../../data/transfer_functions/yellowstone_mt_collection.h5
CPU times: user 8.73 s, sys: 257 ms, total: 8.99 s
Wall time: 9.05 s
for station_id in ["YNP44S", "YNP33B", "WYYS3"]:
mt_data.remove_station(station_id)periods = mt_data.get_periods()interp_periods = np.array([0.1, 1., 10, 100])
interp_mt_data = mt_data.interpolate(interp_periods, inplace=False)1./interp_periodsarray([10. , 1. , 0.1 , 0.01])interp_mt_data.compute_model_errors()
interp_mt_data.model_epsg = 32612
interp_mt_data.compute_relative_locations()sdf = interp_mt_data.to_dataframe()gdf = interp_mt_data.to_geo_df()rx_loc = np.hstack(
(mkvc(gdf.model_north.to_numpy(), 2),
mkvc(gdf.model_east.to_numpy(), 2),
np.zeros((np.prod(gdf.shape[0]), 1)))
)#frequencies = np.array([1e-1, 2])
#station_spacing = 8000
#factor_spacing = 4
#rx_x, rx_y = np.meshgrid(np.arange(0, 50000, station_spacing), np.arange(0, 50000, station_spacing))
#rx_loc = np.hstack((mkvc(rx_x, 2), mkvc(rx_y, 2), np.zeros((np.prod(rx_x.shape), 1))))
#print(rx_loc.shape)
fig, ax = plt.subplots(1,1, figsize=(5,5))
ax.plot(rx_loc[:, 0], rx_loc[:, 1], '.')
ax.set_aspect(1)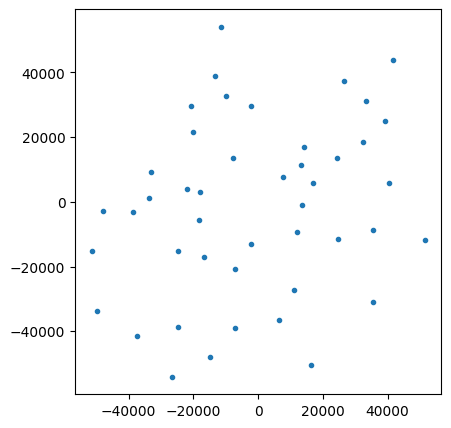
import discretize.utils as dis_utils
from discretize import TreeMesh
from geoana.em.fdem import skin_depth
def get_octree_mesh(
rx_loc, frequencies, sigma_background, station_spacing,
factor_x_pad=2,
factor_y_pad=2,
factor_z_pad_down=2,
factor_z_core=1,
factor_z_pad_up=1,
factor_spacing=4,
):
f_min = frequencies.min()
f_max = frequencies.max()
lx_pad = skin_depth(f_min, sigma_background) * factor_x_pad
ly_pad = skin_depth(f_min, sigma_background) * factor_y_pad
lz_pad_down = skin_depth(f_min, sigma_background) * factor_z_pad_down
lz_core = skin_depth(f_min, sigma_background) * factor_z_core
lz_pad_up = skin_depth(f_min, sigma_background) * factor_z_pad_up
lx_core = rx_loc[:,0].max() - rx_loc[:,0].min()
ly_core = rx_loc[:,1].max() - rx_loc[:,1].min()
lx = lx_pad + lx_core + lx_pad
ly = ly_pad + ly_core + ly_pad
lz = lz_pad_down + lz_core + lz_pad_up
dx = station_spacing / factor_spacing
dy = station_spacing / factor_spacing
dz = np.round(skin_depth(f_max, sigma_background)/4, decimals=-1)
# Compute number of base mesh cells required in x and y
nbcx = 2 ** int(np.ceil(np.log(lx / dx) / np.log(2.0)))
nbcy = 2 ** int(np.ceil(np.log(ly / dy) / np.log(2.0)))
nbcz = 2 ** int(np.ceil(np.log(lz / dz) / np.log(2.0)))
print (dx, dy, dz)
mesh = dis_utils.mesh_builder_xyz(
rx_loc,
[dx, dy, dz],
padding_distance=[[lx_pad, lx_pad], [ly_pad, ly_pad], [lz_pad_down, lz_pad_up]],
depth_core=lz_core,
mesh_type='tree'
)
X, Y = np.meshgrid(mesh.nodes_x, mesh.nodes_y)
topo = np.c_[X.flatten(), Y.flatten(), np.zeros(X.size)]
mesh = refine_tree_xyz(
mesh, topo, octree_levels=[0, 1, 4, 4, 4], method="surface", finalize=False
)
mesh = refine_tree_xyz(
mesh, rx_loc, octree_levels=[1, 0, 0], method="radial", finalize=True
)
return meshmesh = get_octree_mesh(
rx_loc,
1./interp_periods,
1e-2,
5000,
factor_spacing=2,
factor_x_pad=4,
factor_y_pad=4
)
print(mesh.n_cells)2500.0 2500.0 400.0
79459
sigma_background = np.ones(mesh.nC) * 1e-8
ind_active = mesh.cell_centers[:,2] < 0.
sigma_background[ind_active] = 1e-2fig = plt.figure(figsize=(10, 10))
ax = fig.add_subplot(1,2,1)
mesh.plot_slice(
sigma_background, grid=True, normal='Z', ax=ax,
pcolor_opts={"cmap":"turbo", "norm":LogNorm(vmin=1e-3, vmax=1)},
grid_opts={"lw":0.1, "color":'k'},
range_x=(rx_loc[:,0].min()-5000, rx_loc[:,0].max()+5000),
range_y=(rx_loc[:,0].min()-5000, rx_loc[:,0].max()+5000),
slice_loc=0.
)
ax.plot(rx_loc[:,0], rx_loc[:,1], 'ro')
ax.set_aspect(1)
ax2 = fig.add_subplot(1,2,2, sharex=ax)
mesh.plot_slice(
sigma_background, grid=True, normal='X', ax=ax2,
pcolor_opts={"cmap":"turbo", "norm":LogNorm(vmin=1e-4, vmax=10)},
grid_opts={"lw":0.1, "color":'k'},
range_x=(rx_loc[:,0].min()-10000, rx_loc[:,0].max()+10000),
range_y=(-50000, 50000*0.1)
)
ax2.plot(rx_loc[:,0], rx_loc[:,2], 'ro')
ax2.set_aspect(3)
fig.tight_layout()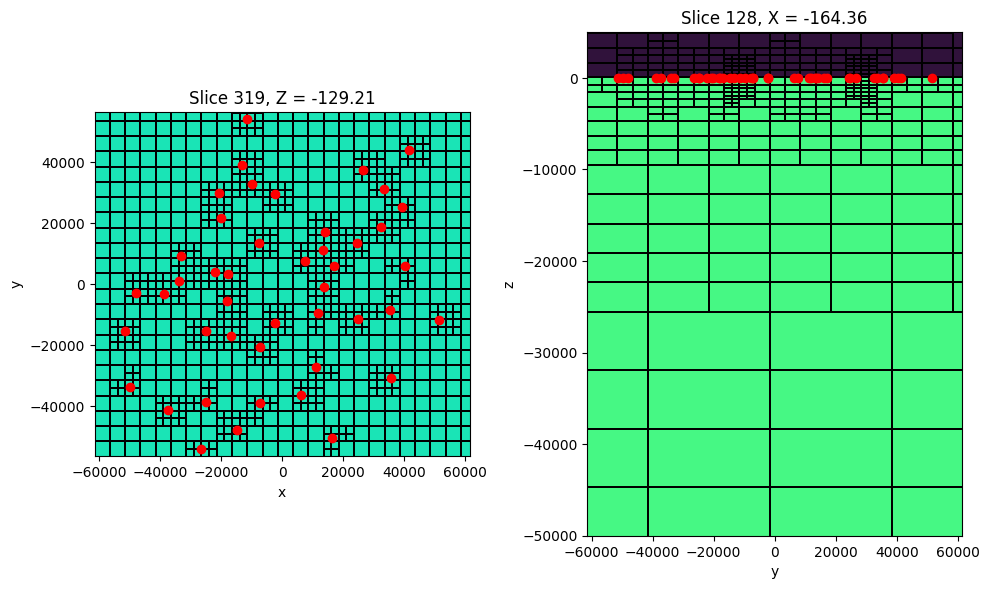
# Generate a Survey
rx_list = []
# rx_orientations_impedance = ['xy', 'yx', 'xx', 'yy']
rx_orientations_impedance = ['xy', 'yx']
for rx_orientation in rx_orientations_impedance:
rx_list.append(
nsem.receivers.PointNaturalSource(
rx_loc, orientation=rx_orientation, component="real"
)
)
rx_list.append(
nsem.receivers.PointNaturalSource(
rx_loc, orientation=rx_orientation, component="imag"
)
)
# rx_orientations_tipper = ['zx', 'zy']
# for rx_orientation in rx_orientations_tipper:
# rx_list.append(
# nsem.receivers.Point3DTipper(
# rx_loc, orientation=rx_orientation, component="real"
# )
# )
# rx_list.append(
# nsem.receivers.Point3DTipper(
# rx_loc, orientation=rx_orientation, component="imag"
# )
# )
# Source list
src_list = [nsem.sources.PlanewaveXYPrimary(rx_list, frequency=f) for f in 1./interp_periods]
# Survey MT
survey = nsem.Survey(src_list)
# rx_orientations = rx_orientations_impedance + rx_orientations_tipper
rx_orientations = rx_orientations_impedance # forward model to get the model response for the synthetic data
# Set the mapping
active_map = maps.InjectActiveCells(
mesh=mesh, indActive=ind_active, valInactive=np.log(1e-8)
)
mapping = maps.ExpMap(mesh) * active_map
# True model
m_true = np.log(sigma_background[ind_active])
# Setup the problem object
simulation = nsem.simulation.Simulation3DPrimarySecondary(
mesh,
survey=survey,
sigmaMap=mapping,
sigmaPrimary=sigma_background,
solver=Solver
)
dpred = simulation.dpred(m_true)constant = 1./np.pi * np.sqrt(5./8.) * 10**3.5print (constant)795.7747154594769
simpeg [zxy, zyx, zxx, zyy, tzx, tzy]
mtpy [zyx, zxy, zyy, zxx, -tzy, -tzx]
frequencies = 1/interp_periods
# components = ["z_xx", "z_xy", "z_yx", "z_yy", "t_zx", "t_zy"]
# components = ["z_yx", "z_xy", "z_yy", "z_xx", "t_zy", "t_zx"]
# components = ["res_yx", "res_xy"]
# components = ["z_yx", "z_xy", "z_yy", "z_xx"]
components = ["z_yx", "z_xy"]
n_rx = rx_loc.shape[0]
n_freq = len(interp_periods)
n_component = 2
n_orientation = len(rx_orientations)
f_dict = dict([(round(ff, 5), ii) for ii, ff in enumerate(1/interp_periods)])
observations = np.zeros((n_freq, n_orientation, n_rx, n_component))
errors = np.zeros_like(observations)
for s_index, station in enumerate(gdf.station):
station_df = sdf.loc[sdf.station == station]
station_df.set_index("period", inplace=True)
for row in station_df.itertuples():
f_index = f_dict[round(1./row.Index, 5)]
for c_index, comp in enumerate(components):
value = getattr(row, comp)
# print (round(1./row.Index, 5), comp, value)
if 't' in comp:
value *= -1
err = getattr(row, f"{comp}_model_error") # user_set error
observations[f_index, c_index, s_index, 0] = value.real
observations[f_index, c_index, s_index, 1] = value.imag
errors[f_index, c_index, s_index, 0] = err
errors[f_index, c_index, s_index, 1] = err
observations[np.where(observations == 0)] = 1e-3
observations[np.isnan(observations)] = 1e-3
# errors = abs(observations) * 0.05
errors[np.where(observations == 1e-3)] = np.infDOBS = dpred.reshape((n_freq, n_orientation, n_component, n_rx))
DCLEAN = DOBS.copy()
FLOOR = np.zeros_like(DOBS)
FLOOR[:,2,0,:] = np.percentile(abs(DOBS[:,2,0,:].flatten()), 90) * 0.1
FLOOR[:,2,1,:] = np.percentile(abs(DOBS[:,2,1,:].flatten()), 90) * 0.1
FLOOR[:,3,0,:] = np.percentile(abs(DOBS[:,3,0,:].flatten()), 90) * 0.1
FLOOR[:,3,1,:] = np.percentile(abs(DOBS[:,3,1,:].flatten()), 90) * 0.1
FLOOR[:,4,0,:] = np.percentile(abs(DOBS[:,4,0,:].flatten()), 90) * 0.1
FLOOR[:,4,1,:] = np.percentile(abs(DOBS[:,4,1,:].flatten()), 90) * 0.1
FLOOR[:,5,0,:] = np.percentile(abs(DOBS[:,5,0,:].flatten()), 90) * 0.1
FLOOR[:,5,1,:] = np.percentile(abs(DOBS[:,5,1,:].flatten()), 90) * 0.1
STD = abs(DOBS) * relative_error + FLOOR
# STD = (FLOOR)
standard_deviation = STD.flatten()
dobs = DOBS.flatten()
dobs += abs(dobs) * relative_error * np.random.randn(dobs.size)
DOBS = dobs.reshape((n_freq, n_orientation, n_component, n_rx))
# + standard_deviation * np.random.randn(standard_deviation.size)def foo_data(i_freq, i_orientation, i_component):
fig, ax = plt.subplots(1,1, figsize=(5, 5))
vmin = np.min([observations[i_freq, i_orientation, :, i_component].min()])
vmax = np.max([observations[i_freq, i_orientation, :, i_component].max()])
out1 = utils.plot2Ddata(rx_loc, observations[i_freq, i_orientation, :, i_component], clim=(vmin, vmax), ax=ax, contourOpts={'cmap':'turbo'}, ncontour=20)
ax.plot(rx_loc[:,0], rx_loc[:,1], 'kx')
if i_orientation<4:
transfer_type = "Z"
else:
transfer_type = "T"
ax.set_title("Frequency={:.1e}, {:s}{:s}-{:s}".format(1./interp_periods[i_freq], transfer_type, rx_orientations[i_orientation], components[i_component]))interact(
foo_data,
i_freq=widgets.IntSlider(min=0, max=n_freq-1),
i_orientation=widgets.IntSlider(min=0, max=n_component-1),
i_component=widgets.IntSlider(min=0, max=n_orientation-1),
)Loading...
constant = 1./np.pi * np.sqrt(5./8.) * 10**3.5dobs = observations.flatten() / constant
standard_deviation = errors.flatten() / constant
dobs[np.isnan(dobs)] = 100.
standard_deviation[dobs==100.] = np.inf# Assign uncertainties
# make data object
data_object = data.Data(survey, dobs=dobs, standard_deviation=standard_deviation)# data_object.standard_deviation = stdnew.flatten() # sim.survey.std
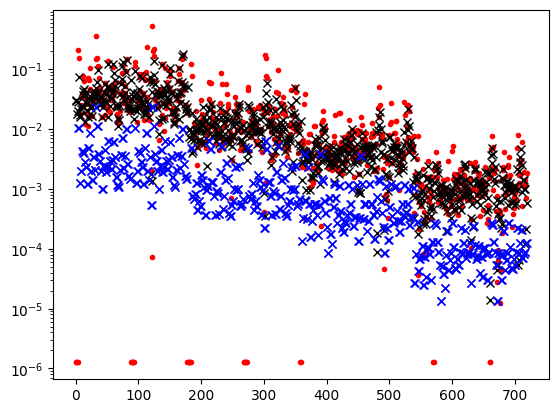
plt.semilogy(abs(data_object.dobs), '.r')
plt.semilogy(abs(data_object.standard_deviation), 'k.', ms=1)
# plt.yscale('symlog')
plt.show()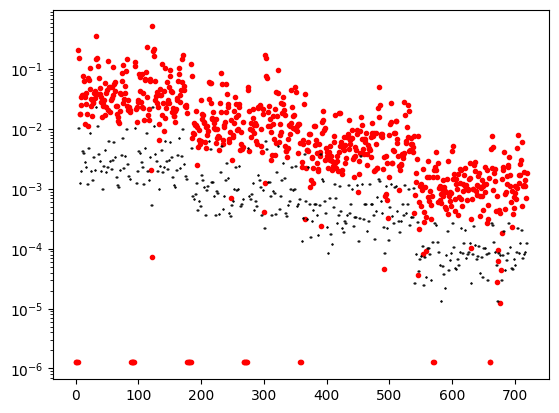
# Set the mapping
# active_map = maps.InjectActiveCells(
# mesh=mesh, indActive=ind_active, valInactive=np.log(1e-8)
# )
# mapping = maps.ExpMap(mesh) * active_map
# True model
m_true = np.log(sigma_background[ind_active])
# # Setup the problem object
# simulation = nsem.simulation.Simulation3DPrimarySecondary(
# mesh,
# survey=survey,
# sigmaMap=mapping,
# sigmaPrimary=sigma_background,
# solver=Solver
# )
mt_sims = []
mt_mappings = []
surveys = []
for i in range(len(src_list)):
# Set the mapping
active_map = maps.InjectActiveCells(
mesh=mesh, indActive=ind_active, valInactive=np.log(1e-8)
)
mapping = maps.ExpMap(mesh) * active_map
survey_chunk = nsem.Survey(src_list[i])
mt_sims.append(
nsem.simulation.Simulation3DPrimarySecondary(
mesh,
survey=survey_chunk,
sigmaMap=maps.IdentityMap(),
sigmaPrimary=sigma_background,
solver=Solver
)
)
surveys.append(survey_chunk)
mt_mappings.append(mapping)
print (f"{i}, {frequencies[i]:.1f} Hz")0, 10.0 Hz
1, 1.0 Hz
2, 0.1 Hz
3, 0.0 Hz
from simpeg.meta import MultiprocessingMetaSimulation
parallel_sim = MultiprocessingMetaSimulation(mt_sims, mt_mappings, n_processes=4)%%time
dpred = parallel_sim.dpred(m_true)Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
CPU times: user 5.88 ms, sys: 3.74 ms, total: 9.62 ms
Wall time: 52.2 s
# data_object.standard_deviation = stdnew.flatten() # sim.survey.std
plt.semilogy(abs(data_object.dobs), '.r')
plt.semilogy(abs(dpred) , 'k.', ms=1)
# plt.yscale('symlog')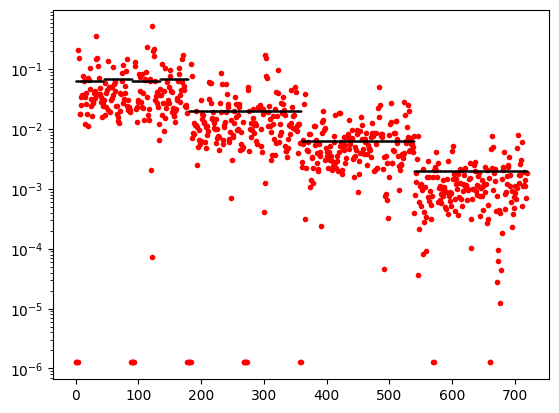
%%time
# Optimization
opt = optimization.ProjectedGNCG(maxIter=10, maxIterCG=20, upper=np.inf, lower=-np.inf)
opt.remember('xc')
# Data misfit
dmis = data_misfit.L2DataMisfit(data=data_object, simulation=parallel_sim)
# Regularization
dz = mesh.h[2].min()
dx = mesh.h[0].min()
regmap = maps.IdentityMap(nP=int(ind_active.sum()))
reg = regularization.Sparse(mesh, active_cells=ind_active, mapping=regmap)
reg.alpha_s = 1e-8
reg.alpha_x = dz/dx
reg.alpha_y = dz/dx
reg.alpha_z = 1.
# Inverse problem
inv_prob = inverse_problem.BaseInvProblem(dmis, reg, opt)
# Beta schedule
beta = directives.BetaSchedule(coolingRate=1, coolingFactor=2)
# Initial estimate of beta
beta_est = directives.BetaEstimate_ByEig(beta0_ratio=1e0)
# Target misfit stop
target = directives.TargetMisfit()
# Create an inversion object
save_dictionary = directives.SaveOutputDictEveryIteration()
directive_list = [beta, beta_est, target, save_dictionary]
inv = inversion.BaseInversion(inv_prob, directiveList=directive_list)
# Set an intitial guess
m_0 = np.log(sigma_background[ind_active])
m_recovered = inv.run(m_0)
Running inversion with SimPEG v0.22.2.dev6+g67b3e9f1c
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 7.86e-06 3.85e+05 0.00e+00 3.85e+05 2.00e+04 0
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
1 3.93e-06 5.30e+04 8.25e+06 5.30e+04 1.98e+03 0
2 1.97e-06 4.24e+04 3.69e+07 4.25e+04 4.05e+03 0 Skip BFGS
3 9.83e-07 3.81e+04 4.81e+07 3.81e+04 2.14e+03 0
4 4.91e-07 3.14e+04 5.82e+07 3.14e+04 1.74e+03 0
5 2.46e-07 2.68e+04 7.22e+07 2.69e+04 5.55e+02 0
6 1.23e-07 2.66e+04 9.58e+07 2.66e+04 6.74e+02 0
7 6.14e-08 2.55e+04 1.05e+08 2.55e+04 4.56e+02 0
8 3.07e-08 2.48e+04 1.35e+08 2.49e+04 4.46e+02 0
9 1.54e-08 2.39e+04 1.35e+08 2.39e+04 2.71e+02 1
10 7.68e-09 2.39e+04 1.74e+08 2.39e+04 4.48e+02 0
------------------------- STOP! -------------------------
1 : |fc-fOld| = 1.8631e+00 <= tolF*(1+|f0|) = 3.8477e+04
1 : |xc-x_last| = 4.8178e+01 <= tolX*(1+|x0|) = 5.5358e+01
0 : |proj(x-g)-x| = 4.4798e+02 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 4.4798e+02 <= 1e3*eps = 1.0000e-02
1 : maxIter = 10 <= iter = 10
------------------------- DONE! -------------------------
CPU times: user 41.8 s, sys: 1min 52s, total: 2min 34s
Wall time: 13min 35s
iteration = len(save_dictionary.outDict)
iteration = 2
m = save_dictionary.outDict[iteration]['m']
sigma_est = mapping*m
pred = save_dictionary.outDict[iteration]['dpred']
# DPRED = pred.reshape((n_freq, n_orientation, n_component, n_rx))
# DOBS = dpred.reshape((n_freq, n_orientation, n_component, n_rx))
# MISFIT = (DPRED-DOBS)/STD
# data_object.standard_deviation = stdnew.flatten() # sim.survey.std
plt.semilogy(abs(data_object.dobs), '.r')
plt.semilogy(abs(pred) , 'kx')
plt.semilogy(abs(standard_deviation) , 'bx')
# plt.yscale('symlog')
# plt.semilogy(abs(data_object.standard_deviation), 'b.', ms=1)
plt.show()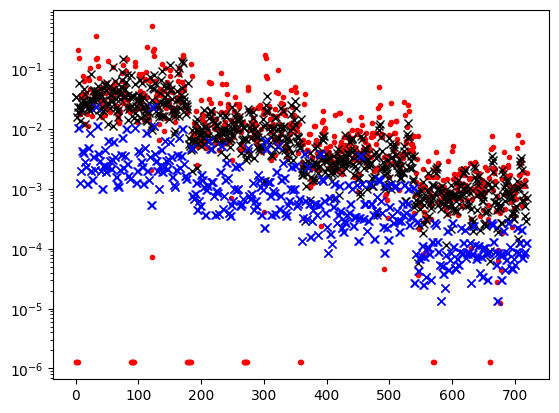
fig, ax = plt.subplots(1,1, figsize=(5, 5))
z_loc = -0.
y_loc = 0.
mesh.plot_slice(
sigma_est, grid=True, normal='Z', ax=ax,
pcolor_opts={"cmap":"turbo", "norm":LogNorm(vmin=5e-3, vmax=1e-1)},
range_x=(rx_loc[:,0].min()-10000, rx_loc[:,0].max()+10000),
range_y=(rx_loc[:,0].min()-10000, rx_loc[:,0].max()+10000),
slice_loc=z_loc
)
ax.set_yticklabels([])
ax.plot(rx_loc[:,0], rx_loc[:,1], 'kx')
ax.set_aspect(1)
ax.set_xlabel("Easting (m)")
ax.set_ylabel("Northing (m)")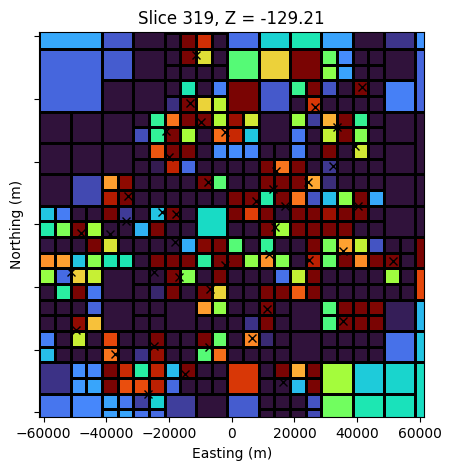
fig, ax = plt.subplots(1,1, figsize=(5, 5))
mesh.plot_slice(
sigma_est, grid=False, normal='Y', ax=ax,
pcolor_opts={"cmap":"turbo", "norm":LogNorm(vmin=2e-3, vmax=1e-1)},
range_x=(rx_loc[:,0].min()-10000, rx_loc[:,0].max()+10000),
# range_y=(-lx_core, lx_core*0.1),
slice_loc=y_loc
)
# ax.set_yticklabels([])
# ax.plot(rx_loc[:,0], rx_loc[:,2], 'kx')
ax.set_aspect(0.54)
ax.set_xlabel("Easting (m)")
ax.set_ylabel("Elevation (m)")
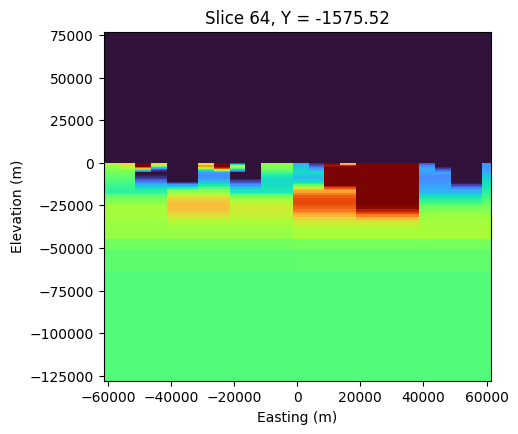
from ipywidgets import interact, widgets# def foo_misfit(i_freq, i_orientation, i_component):
# fig, axs = plt.subplots(1,3, figsize=(15, 5))
# ax1, ax2, ax3 = axs
# vmin = np.min([DOBS[i_freq, i_orientation, i_component,:].min(), DPRED[i_freq, i_orientation, i_component,:].min()])
# vmax = np.max([DOBS[i_freq, i_orientation, i_component,:].max(), DPRED[i_freq, i_orientation, i_component,:].max()])
# out1 = utils.plot2Ddata(rx_loc, DOBS[i_freq, i_orientation, i_component,:], clim=(vmin, vmax), ax=ax1, contourOpts={'cmap':'turbo'}, ncontour=20)
# out2 = utils.plot2Ddata(rx_loc, DPRED[i_freq, i_orientation, i_component,:], clim=(vmin, vmax), ax=ax2, contourOpts={'cmap':'turbo'}, ncontour=20)
# out3 = utils.plot2Ddata(rx_loc, MISFIT[i_freq, i_orientation, i_component,:], clim=(-5, 5), ax=ax3, contourOpts={'cmap':'turbo'}, ncontour=20)
# ax.plot(rx_loc[:,0], rx_loc[:,1], 'kx')# interact(
# foo_misfit,
# i_freq=widgets.IntSlider(min=0, max=n_freq-1),
# i_orientation=widgets.IntSlider(min=0, max=n_orientation-1),
# i_component=widgets.IntSlider(min=0, max=n_component-1),
# )models = {}
models["estimated_sigma"] = sigma_est
mesh.write_vtk("yellowstone",models=models)