Note: this example assumes that data availability (Network, Station, Channel, Start, End) are all previously known. If you do not know the data that you want to download use IRIS tools to get data availability.
from pathlib import Path
import numpy as np
import pandas as pd
from mth5.mth5 import MTH5
from mth5.clients.fdsn import FDSN
from mth5.clients.make_mth5 import MakeMTH5
from matplotlib import pyplot as plt
%matplotlib inlineSet the path to save files to as the current working directory¶
default_path = Path().cwd()
print(default_path)c:\Users\jpeacock\OneDrive - DOI\Documents\GitHub\iris-mt-course-2022\notebooks\mth5
Initialize a MakeMTH5 object¶
Here, we are setting the MTH5 file version to 0.2.0 so that we can have multiple surveys in a single file. Also, setting the client to “IRIS”. Here, we are using obspy.clients tools for the request. Here are the available FDSN clients.
Note: Only the “IRIS” client has been tested.
# FDSN object is used for instructional purposes
fdsn_object = FDSN(mth5_version='0.2.0')
fdsn_object.client = "IRIS"
Make the data inquiry as a DataFrame¶
There are a few ways to make the inquiry to request data.
Make a DataFrame by hand. Here we will make a list of entries and then create a DataFrame with the proper column names
You can create a CSV file with a row for each entry. There are some formatting that you need to be aware of. That is the column names and making sure that date-times are YYYY-MM-DDThh:mm:ss
| Column Name | Description |
|---|---|
| network | FDSN Network code (2 letters) |
| station | FDSN Station code (usually 5 characters) |
| location | FDSN Location code (typically not used for MT) |
| channel | FDSN Channel code (3 characters) |
| start | Start time (YYYY-MM-DDThh:mm:ss) UTC |
| end | End time (YYYY-MM-DDThh:mm:ss) UTC |
From IRIS Metadata¶
Here are the station running during WYYS2, the original transfer function was processed with MTF20.
| Station | Start | End |
|---|---|---|
| MTF18 | 2009-07-11T00:07:55.0000 | 2009-08-28T21:31:41.0000 |
| MTF19 | 2009-07-10T00:17:16.0000 | 2009-09-16T19:39:22.0000 |
| MTF20 | 2009-07-03T22:38:13.0000 | 2009-08-13T17:15:39.0000 |
| MTF21 | 2009-07-31T21:45:22.0000 | 2009-09-18T23:56:11.0000 |
| WYI17 | 2009-07-13T22:17:15.0000 | 2009-08-04T19:41:33.0000 |
| WYI21 | 2009-07-27T07:26:00.0000 | 2009-09-11T16:36:30.0000 |
| WYJ18 | 2009-07-13T00:58:30.0000 | 2009-08-03T19:39:24.0000 |
| WYJ21 | 2009-07-23T22:23:20.0000 | 2009-08-16T21:03:53.0000 |
| WYK21 | 2009-07-22T22:01:44.0000 | 2009-08-06T12:11:52.0000 |
| WYL21 | 2009-07-21T20:27:41.0000 | 2009-08-18T17:56:39.0000 |
| WYYS1 | 2009-07-14T21:58:53.0000 | 2009-08-05T21:14:56.0000 |
| WYYS2 | 2009-07-15T22:43:28.0000 | 2009-08-20T00:17:06.0000 |
Here are the station running during WYYS3, the original transfer function was processed with MTC18
| Station | Start | End |
|---|---|---|
| MTC18 | 2009-08-21T21:52:53.0000 | 2009-09-13T00:02:18.0000 |
| MTE18 | 2009-08-07T20:40:38.0000 | 2009-08-28T19:03:01.0000 |
| MTE19 | 2009-08-12T22:45:47.0000 | 2009-09-02T17:10:53.0000 |
| MTE20 | 2009-08-10T21:12:58.0000 | 2009-09-19T20:25:30.0000 |
| MTE21 | 2009-08-09T02:45:02.0000 | 2009-08-30T21:43:43.0000 |
| WYH21 | 2009-08-01T22:50:53.0000 | 2009-08-24T20:23:03.0000 |
| WYG21 | 2009-08-03T02:56:34.0000 | 2009-08-18T19:56:53.0000 |
| WYYS3 | 2009-08-20T01:55:41.0000 | 2009-09-17T20:04:21.0000 |
In the examples below we will just download the original remote references, but as an excersize you could add in some other stations.
channels = ["LFE", "LFN", "LFZ", "LQE", "LQN"]
WYYS2 = ["4P", "WYYS2", "2009-07-15T00:00:00", "2009-08-21T00:00:00"]
MTF20 = ["4P", "MTF20", "2009-07-03T00:00:00", "2009-08-13T00:00:00"]
WYYS3 = ["4P", "WYYS3", "2009-08-20T00:00:00", "2009-09-17T00:00:00"]
MTC18 = ["4P", "MTC20", "2009-08-21T00:00:00", "2009-09-13T00:00:00"]
request_list = []
for entry in [WYYS2, MTF20, WYYS3, MTC18]:
for channel in channels:
request_list.append(
[entry[0], entry[1], "", channel, entry[2], entry[3]]
)
# Turn list into dataframe
request_df = pd.DataFrame(request_list, columns=fdsn_object.request_columns)
request_dfSave the request as a CSV¶
Its helpful to be able to save the request as a CSV and modify it and use it later. A CSV can be input as a request to MakeMTH5
request_df.to_csv(default_path.joinpath("fdsn_request.csv"))Get only the metadata from IRIS¶
It can be helpful to make sure that your request is what you would expect. For that you can request only the metadata from IRIS. The request is quick and light so shouldn’t need to worry about the speed. This returns a StationXML file and is loaded into an obspy.Inventory object.
inventory, data = fdsn_object.get_inventory_from_df(request_df, data=False)/opt/conda/lib/python3.10/site-packages/mth5/clients/fdsn.py:550: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
sub_df.drop_duplicates("station", inplace=True)
/opt/conda/lib/python3.10/site-packages/mth5/clients/fdsn.py:550: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
sub_df.drop_duplicates("station", inplace=True)
/opt/conda/lib/python3.10/site-packages/mth5/clients/fdsn.py:550: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
sub_df.drop_duplicates("station", inplace=True)
/opt/conda/lib/python3.10/site-packages/mth5/clients/fdsn.py:550: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
sub_df.drop_duplicates("station", inplace=True)
Have a look at the Inventory to make sure it contains what is requested.
inventory.write(default_path.joinpath("yellowstone_stationxml.xml").as_posix(), format="stationxml")
print(inventory)Inventory created at 2024-09-20T00:40:31.940624Z
Created by: ObsPy 1.4.1
https://www.obspy.org
Sending institution: MTH5
Contains:
Networks (4):
4P (4x)
Stations (4):
4P.MTC20 (Zortman, MT, USA)
4P.MTF20 (Billings, MT, USA)
4P.WYYS2 (Grant Village, WY, USA)
4P.WYYS3 (Fishing Bridge, WY, USA)
Channels (26):
4P.MTC20..LFZ, 4P.MTC20..LFN, 4P.MTC20..LFE, 4P.MTC20..LQN,
4P.MTC20..LQE, 4P.MTF20..LFZ, 4P.MTF20..LFN (2x),
4P.MTF20..LFE (2x), 4P.MTF20..LQN (2x), 4P.MTF20..LQE (2x),
4P.WYYS2..LFZ, 4P.WYYS2..LFN, 4P.WYYS2..LFE, 4P.WYYS2..LQN,
4P.WYYS2..LQE, 4P.WYYS3..LFZ, 4P.WYYS3..LFN, 4P.WYYS3..LFE,
4P.WYYS3..LQN (2x), 4P.WYYS3..LQE (2x)
Make an MTH5 from a request¶
Now that we’ve created a request, and made sure that its what we expect, we can make an MTH5 file. The input can be either the DataFrame or the CSV file.
Here we are going to use the MakeMTH5 object to create from an FDSN client.
We are going to time it just to get an indication how long it might take. Should take a couple of minutes depending on connection.
%%time
mth5_object = MakeMTH5.from_fdsn_client(request_df, interact=False)
print(f"Created {mth5_object}")2024-10-14T14:35:37.707606-0700 | INFO | mth5.mth5 | _initialize_file | Initialized MTH5 0.2.0 file c:\Users\jpeacock\OneDrive - DOI\Documents\GitHub\iris-mt-course-2022\notebooks\mth5\4P_WYYS2_MTF20_WYYS3_MTC20.h5 in mode w
C:\Users\jpeacock\OneDrive - DOI\Documents\GitHub\mth5\mth5\clients\fdsn.py:561: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
sub_df.drop_duplicates("station", inplace=True)
C:\Users\jpeacock\OneDrive - DOI\Documents\GitHub\mth5\mth5\clients\fdsn.py:561: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
sub_df.drop_duplicates("station", inplace=True)
C:\Users\jpeacock\OneDrive - DOI\Documents\GitHub\mth5\mth5\clients\fdsn.py:561: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
sub_df.drop_duplicates("station", inplace=True)
C:\Users\jpeacock\OneDrive - DOI\Documents\GitHub\mth5\mth5\clients\fdsn.py:561: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
sub_df.drop_duplicates("station", inplace=True)
2024-10-14T14:39:35.090107-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_si_units to a CoefficientFilter.
2024-10-14T14:39:35.131113-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_dipole_100.000 to a CoefficientFilter.
2024-10-14T14:39:35.375772-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_si_units to a CoefficientFilter.
2024-10-14T14:39:35.415768-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_dipole_100.000 to a CoefficientFilter.
2024-10-14T14:39:36.030731-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_si_units to a CoefficientFilter.
2024-10-14T14:39:36.052718-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_dipole_100.000 to a CoefficientFilter.
2024-10-14T14:39:36.271433-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_si_units to a CoefficientFilter.
2024-10-14T14:39:36.295407-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_dipole_100.000 to a CoefficientFilter.
2024-10-14T14:39:36.419984-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_si_units to a CoefficientFilter.
2024-10-14T14:39:36.441953-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_dipole_100.000 to a CoefficientFilter.
2024-10-14T14:39:36.570065-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_si_units to a CoefficientFilter.
2024-10-14T14:39:36.592066-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_dipole_100.000 to a CoefficientFilter.
2024-10-14T14:39:36.987415-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_si_units to a CoefficientFilter.
2024-10-14T14:39:37.012386-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_dipole_100.000 to a CoefficientFilter.
2024-10-14T14:39:37.128988-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_si_units to a CoefficientFilter.
2024-10-14T14:39:37.149030-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_dipole_100.000 to a CoefficientFilter.
2024-10-14T14:39:37.264107-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_si_units to a CoefficientFilter.
2024-10-14T14:39:37.286105-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_dipole_100.000 to a CoefficientFilter.
2024-10-14T14:39:37.406667-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_si_units to a CoefficientFilter.
2024-10-14T14:39:37.428670-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_dipole_100.000 to a CoefficientFilter.
2024-10-14T14:39:37.781985-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_si_units to a CoefficientFilter.
2024-10-14T14:39:37.800992-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_dipole_100.000 to a CoefficientFilter.
2024-10-14T14:39:37.917620-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_si_units to a CoefficientFilter.
2024-10-14T14:39:37.946652-0700 | INFO | mt_metadata.timeseries.filters.obspy_stages | create_filter_from_stage | Converting PoleZerosResponseStage electric_dipole_100.000 to a CoefficientFilter.
2024-10-14T14:39:45.213007-0700 | INFO | mth5.groups.base | _add_group | RunGroup a already exists, returning existing group.
2024-10-14T14:39:46.428171-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:39:46.994506-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:39:47.596312-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:39:48.114192-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:39:48.627996-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:39:48.811049-0700 | INFO | mth5.groups.base | _add_group | RunGroup b already exists, returning existing group.
2024-10-14T14:39:51.551963-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:39:52.709159-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:39:54.260964-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:39:55.554133-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:39:56.815276-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:39:57.240232-0700 | INFO | mth5.groups.base | _add_group | RunGroup c already exists, returning existing group.
2024-10-14T14:40:01.564901-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:02.786256-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:04.041310-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:04.962234-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:05.879616-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:06.277253-0700 | INFO | mth5.groups.base | _add_group | RunGroup a already exists, returning existing group.
2024-10-14T14:40:08.121777-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:40:09.191976-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:40:10.103378-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:40:11.422186-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:40:13.324134-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:40:13.882025-0700 | INFO | mth5.groups.base | _add_group | RunGroup b already exists, returning existing group.
2024-10-14T14:40:21.633475-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:40:22.403808-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:40:23.344515-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:40:24.168872-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:40:24.913391-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:40:25.213407-0700 | INFO | mth5.groups.base | _add_group | RunGroup c already exists, returning existing group.
2024-10-14T14:40:27.240273-0700 | WARNING | mth5.timeseries.run_ts | validate_metadata | end time of dataset 2009-08-13T00:00:00+00:00 does not match metadata end 2009-08-13T17:15:39+00:00 updating metatdata value to 2009-08-13T00:00:00+00:00
2024-10-14T14:40:27.773350-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:28.285873-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:28.815818-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:29.396923-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:30.020015-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:30.339637-0700 | WARNING | mth5.clients.fdsn | wrangle_runs_into_containers | More or less runs have been requested by the user than are defined in the metadata. Runs will be defined but only the requested run extents contain time series data based on the users request.
2024-10-14T14:40:30.384655-0700 | INFO | mth5.groups.base | _add_group | RunGroup c already exists, returning existing group.
2024-10-14T14:40:32.673915-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:33.360057-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:33.955432-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:34.635047-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:35.336019-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:35.604091-0700 | INFO | mth5.groups.base | _add_group | RunGroup d already exists, returning existing group.
2024-10-14T14:40:35.738200-0700 | INFO | mth5.groups.base | _add_group | RunGroup c already exists, returning existing group.
2024-10-14T14:40:35.858729-0700 | INFO | mth5.groups.base | _add_group | RunGroup d already exists, returning existing group.
2024-10-14T14:40:37.740960-0700 | WARNING | mth5.timeseries.run_ts | validate_metadata | end time of dataset 2009-09-09T20:39:00+00:00 does not match metadata end 2009-09-17T20:04:21+00:00 updating metatdata value to 2009-09-09T20:39:00+00:00
2024-10-14T14:40:38.244832-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id d. Setting to ch.run_metadata.id to d
2024-10-14T14:40:38.747770-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id d. Setting to ch.run_metadata.id to d
2024-10-14T14:40:39.242730-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id d. Setting to ch.run_metadata.id to d
2024-10-14T14:40:39.675924-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id d. Setting to ch.run_metadata.id to d
2024-10-14T14:40:40.131108-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id d. Setting to ch.run_metadata.id to d
2024-10-14T14:40:40.311614-0700 | INFO | mth5.groups.base | _add_group | RunGroup a already exists, returning existing group.
2024-10-14T14:40:41.062867-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:40:41.512353-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:40:41.941612-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:40:42.371216-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:40:42.835527-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id a. Setting to ch.run_metadata.id to a
2024-10-14T14:40:42.994715-0700 | INFO | mth5.groups.base | _add_group | RunGroup b already exists, returning existing group.
2024-10-14T14:40:44.450525-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:40:44.971342-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:40:45.562739-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:40:46.475546-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:40:47.439380-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id b. Setting to ch.run_metadata.id to b
2024-10-14T14:40:47.827674-0700 | INFO | mth5.groups.base | _add_group | RunGroup c already exists, returning existing group.
2024-10-14T14:40:49.882952-0700 | WARNING | mth5.timeseries.run_ts | validate_metadata | end time of dataset 2009-09-13T00:00:00+00:00 does not match metadata end 2009-09-14T22:50:17+00:00 updating metatdata value to 2009-09-13T00:00:00+00:00
2024-10-14T14:40:50.382123-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:50.904544-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:51.445900-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:52.048900-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:52.557388-0700 | WARNING | mth5.groups.run | from_runts | Channel run.id sr1_001 != group run.id c. Setting to ch.run_metadata.id to c
2024-10-14T14:40:53.371604-0700 | INFO | mth5.mth5 | close_mth5 | Flushing and closing c:\Users\jpeacock\OneDrive - DOI\Documents\GitHub\iris-mt-course-2022\notebooks\mth5\4P_WYYS2_MTF20_WYYS3_MTC20.h5
2024-10-14T14:40:53.375572-0700 | WARNING | mth5.mth5 | filename | MTH5 file is not open or has not been created yet. Returning default name
Created c:\Users\jpeacock\OneDrive - DOI\Documents\GitHub\iris-mt-course-2022\notebooks\mth5\4P_WYYS2_MTF20_WYYS3_MTC20.h5
CPU times: total: 40.5 s
Wall time: 5min 16s
# open file already created
mth5_object = MTH5()
mth5_object = mth5_object.open_mth5("4P_WYYS2_MTF20_WYYS3_MTC20.h5")Have a look at the contents of the created file¶
mth5_object/:
====================
|- Group: Experiment
--------------------
|- Group: Reports
-----------------
|- Group: Standards
-------------------
--> Dataset: summary
......................
|- Group: Surveys
-----------------
|- Group: 4P
------------
|- Group: Filters
-----------------
|- Group: coefficient
---------------------
|- Group: electric_analog_to_digital
------------------------------------
|- Group: electric_dipole_100.000
---------------------------------
|- Group: electric_si_units
---------------------------
|- Group: magnetic_analog_to_digital
------------------------------------
|- Group: fap
-------------
|- Group: fir
-------------
|- Group: time_delay
--------------------
|- Group: electric_time_offset
------------------------------
|- Group: hx_time_offset
------------------------
|- Group: hy_time_offset
------------------------
|- Group: hz_time_offset
------------------------
|- Group: zpk
-------------
|- Group: electric_butterworth_high_pass_6000
---------------------------------------------
--> Dataset: poles
....................
--> Dataset: zeros
....................
|- Group: electric_butterworth_low_pass
---------------------------------------
--> Dataset: poles
....................
--> Dataset: zeros
....................
|- Group: magnetic_butterworth_low_pass
---------------------------------------
--> Dataset: poles
....................
--> Dataset: zeros
....................
|- Group: Reports
-----------------
|- Group: Standards
-------------------
--> Dataset: summary
......................
|- Group: Stations
------------------
|- Group: MTC20
---------------
|- Group: Fourier_Coefficients
------------------------------
|- Group: Transfer_Functions
----------------------------
|- Group: a
-----------
--> Dataset: ex
.................
--> Dataset: ey
.................
--> Dataset: hx
.................
--> Dataset: hy
.................
--> Dataset: hz
.................
|- Group: b
-----------
--> Dataset: ex
.................
--> Dataset: ey
.................
--> Dataset: hx
.................
--> Dataset: hy
.................
--> Dataset: hz
.................
|- Group: c
-----------
--> Dataset: ex
.................
--> Dataset: ey
.................
--> Dataset: hx
.................
--> Dataset: hy
.................
--> Dataset: hz
.................
|- Group: MTF20
---------------
|- Group: Fourier_Coefficients
------------------------------
|- Group: Transfer_Functions
----------------------------
|- Group: a
-----------
--> Dataset: ex
.................
--> Dataset: ey
.................
--> Dataset: hx
.................
--> Dataset: hy
.................
--> Dataset: hz
.................
|- Group: b
-----------
--> Dataset: ex
.................
--> Dataset: ey
.................
--> Dataset: hx
.................
--> Dataset: hy
.................
--> Dataset: hz
.................
|- Group: c
-----------
--> Dataset: ex
.................
--> Dataset: ey
.................
--> Dataset: hx
.................
--> Dataset: hy
.................
--> Dataset: hz
.................
|- Group: WYYS2
---------------
|- Group: Fourier_Coefficients
------------------------------
|- Group: Transfer_Functions
----------------------------
|- Group: a
-----------
--> Dataset: ex
.................
--> Dataset: ey
.................
--> Dataset: hx
.................
--> Dataset: hy
.................
--> Dataset: hz
.................
|- Group: b
-----------
--> Dataset: ex
.................
--> Dataset: ey
.................
--> Dataset: hx
.................
--> Dataset: hy
.................
--> Dataset: hz
.................
|- Group: c
-----------
--> Dataset: ex
.................
--> Dataset: ey
.................
--> Dataset: hx
.................
--> Dataset: hy
.................
--> Dataset: hz
.................
|- Group: WYYS3
---------------
|- Group: Fourier_Coefficients
------------------------------
|- Group: Transfer_Functions
----------------------------
|- Group: c
-----------
--> Dataset: ex
.................
--> Dataset: ey
.................
--> Dataset: hx
.................
--> Dataset: hy
.................
--> Dataset: hz
.................
|- Group: d
-----------
--> Dataset: ex
.................
--> Dataset: ey
.................
--> Dataset: hx
.................
--> Dataset: hy
.................
--> Dataset: hz
.................
--> Dataset: channel_summary
..............................
--> Dataset: fc_summary
.........................
--> Dataset: tf_summary
.........................Channel Summary¶
A convenience table is supplied with an MTH5 file. This table provides some information about each channel that is present in the file. It also provides columns hdf5_reference, run_hdf5_reference, and station_hdf5_reference, these are internal references within an HDF5 file and can be used to directly access a group or dataset by using mth5_object.from_reference method.
Note: When a MTH5 file is close the table is resummarized so when you open the file next the channel_summary will be up to date. Same with the tf_summary.
mth5_object.channel_summary.clear_table()
mth5_object.channel_summary.summarize()
ch_df = mth5_object.channel_summary.to_dataframe()
ch_dfRun Summary¶
Have a look at the run summary.
mth5_object.run_summaryHave a look at a station¶
Lets grab one station CAS04 and have a look at its metadata and contents.
Here we will grab it from the mth5_object.
wyys2 = mth5_object.get_station("WYYS2", survey="4P")
wyys2.metadata{
"station": {
"acquired_by.name": null,
"channels_recorded": [
"ex",
"ey",
"hx",
"hy",
"hz"
],
"data_type": "MT",
"fdsn.id": "WYYS2",
"geographic_name": "Grant Village, WY, USA",
"hdf5_reference": "<HDF5 object reference>",
"id": "WYYS2",
"location.declination.comments": "igrf.m by Drew Compston",
"location.declination.model": "IGRF-13",
"location.declination.value": 12.5095194946954,
"location.elevation": 2392.334716797,
"location.latitude": 44.39635,
"location.longitude": -110.577,
"mth5_type": "Station",
"orientation.method": "compass",
"orientation.reference_frame": "geographic",
"provenance.archive.name": null,
"provenance.creation_time": "1980-01-01T00:00:00+00:00",
"provenance.creator.name": null,
"provenance.software.author": "Anna Kelbert, USGS",
"provenance.software.name": "mth5_metadata.m",
"provenance.software.version": "2024-06-12",
"provenance.submitter.email": null,
"provenance.submitter.name": null,
"provenance.submitter.organization": null,
"release_license": "CC0-1.0",
"run_list": [
"a",
"b",
"c"
],
"time_period.end": "2009-08-20T00:17:06+00:00",
"time_period.start": "2009-07-15T22:43:28+00:00"
}
}Changing Metadata¶
If you want to change the metadata of any group, be sure to use the write_metadata method. Here’s an example:
wyys2.metadata.location.declination.value = 12.2
wyys2.write_metadata()
print(wyys2.metadata.location.declination)declination:
comments = igrf.m by Drew Compston
model = IGRF-13
value = 12.2
Have a look at a single channel¶
Let’s pick out a channel and interogate it. There are a couple ways
Get a channel the first will be from the
hdf5_reference[demonstrated here]Get a channel from
mth5_objectGet a station first then get a channel
ex = mth5_object.from_reference(ch_df.iloc[50].hdf5_reference).to_channel_ts()
print(ex)Channel Summary:
Survey: 4P
Station: WYYS3
Run: c
Channel Type: Magnetic
Component: hz
Sample Rate: 1.0
Start: 2009-08-20T01:55:41+00:00
End: 2009-08-28T17:23:45+00:00
N Samples: 746885
ex.channel_metadata{
"magnetic": {
"channel_number": 0,
"comments": "run_ids: [d,c]",
"component": "hz",
"data_quality.rating.value": 0,
"filter.applied": [
true,
true,
true
],
"filter.name": [
"magnetic_butterworth_low_pass",
"magnetic_analog_to_digital",
"hz_time_offset"
],
"hdf5_reference": "<HDF5 object reference>",
"location.elevation": 2379.8,
"location.latitude": 44.5607,
"location.longitude": -110.315,
"measurement_azimuth": 0.0,
"measurement_tilt": 90.0,
"mth5_type": "Magnetic",
"sample_rate": 1.0,
"sensor.id": "2612-18",
"sensor.manufacturer": "Barry Narod",
"sensor.model": "fluxgate NIMS",
"sensor.name": "NIMS",
"sensor.type": "Magnetometer",
"time_period.end": "2009-08-28T17:23:45+00:00",
"time_period.start": "2009-08-20T01:55:41+00:00",
"type": "magnetic",
"units": "digital counts"
}
}Calibrate time series data¶
Most data loggers output data in digital counts. Then a series of filters that represent the various instrument responses are applied to get the data into physical units. The data can then be analyzed and processed. Commonly this is done during the processing step, but it is important to be able to look at time series data in physical units. Here we provide a remove_instrument_response method in the ChananelTS object. Here’s an example:
print(ex.channel_response)
ex.channel_response.plot_response(np.logspace(0, 4, 50))Filters Included:
=========================
pole_zero_filter:
calibration_date = 1980-01-01
comments = NIMS magnetic field 3 pole Butterworth 0.5 low pass (analog)
gain = 1.0
name = magnetic_butterworth_low_pass
normalization_factor = 2002.26936395594
poles = [ -6.283185+10.882477j -6.283185-10.882477j -12.566371 +0.j ]
type = zpk
units_in = nT
units_out = V
zeros = []
--------------------
coefficient_filter:
calibration_date = 1980-01-01
comments = analog to digital conversion (magnetic)
gain = 100.0
name = magnetic_analog_to_digital
type = coefficient
units_in = V
units_out = count
--------------------
time_delay_filter:
calibration_date = 1980-01-01
comments = time offset in seconds (digital)
delay = -0.21
gain = 1.0
name = hz_time_offset
type = time delay
units_in = count
units_out = count
--------------------
2024-09-19T17:44:25.205420-0700 | WARNING | mt_metadata.timeseries.filters.channel_response | complex_response | Filters list not provided, building list assuming all are applied
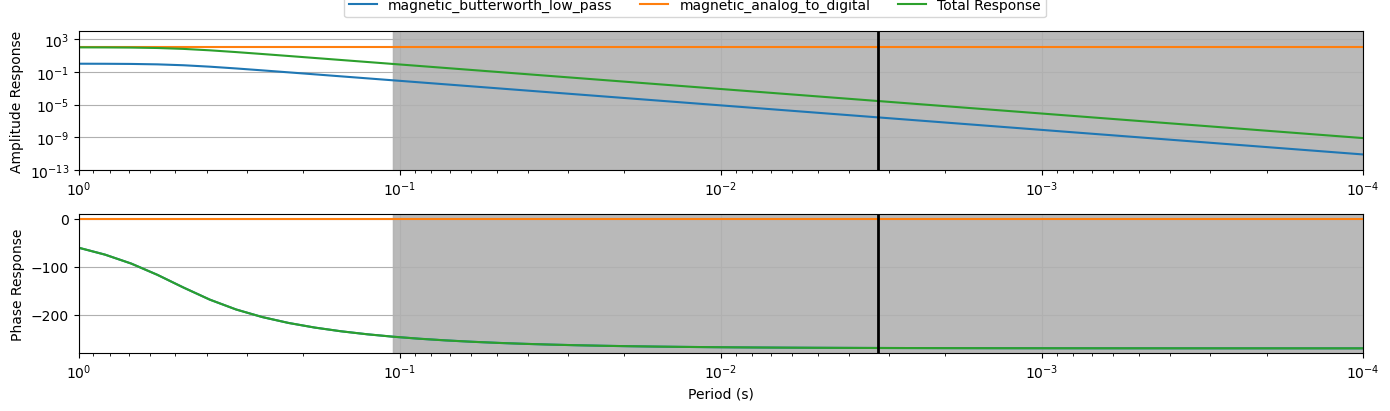
ex.remove_instrument_response(plot=True)/opt/conda/lib/python3.10/site-packages/mth5/timeseries/ts_filters.py:546: UserWarning: Attempt to set non-positive xlim on a log-scaled axis will be ignored.
ax2.set_xlim((f[0], f[-1]))
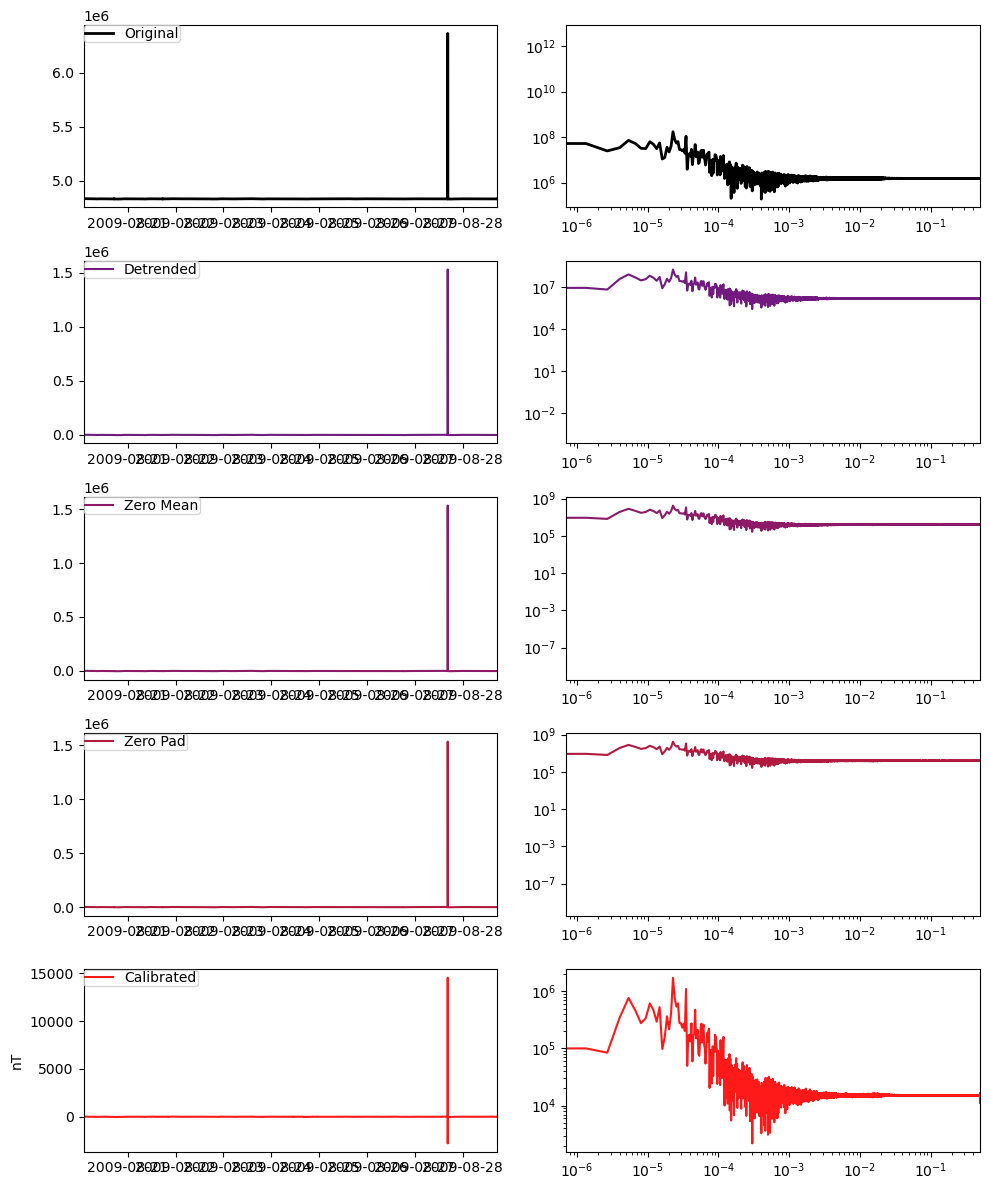
Channel Summary:
Survey: 4P
Station: WYYS3
Run: c
Channel Type: Magnetic
Component: hz
Sample Rate: 1.0
Start: 2009-08-20T01:55:41+00:00
End: 2009-08-28T17:23:45+00:00
N Samples: 746885Have a look at a run¶
Let’s pick out a run, take a slice of it, and interogate it. There are a couple ways
Get a run the first will be from the
run_hdf5_reference[demonstrated here]Get a run from
mth5_objectGet a station first then get a run
run_from_reference = mth5_object.from_reference(ch_df.iloc[0].run_hdf5_reference).to_runts(start=ch_df.iloc[0].start.isoformat(), n_samples=360)
print(run_from_reference)2024-09-19T17:44:31.517989-0700 | WARNING | mth5.timeseries.run_ts | validate_metadata | end time of dataset 2009-08-23T00:00:14+00:00 does not match metadata end 2009-08-23T01:04:36+00:00 updating metatdata value to 2009-08-23T00:00:14+00:00
RunTS Summary:
Survey: 4P
Station: MTC20
Run: a
Start: 2009-08-22T23:54:15+00:00
End: 2009-08-23T00:00:14+00:00
Sample Rate: 1.0
Components: ['ex', 'ey', 'hx', 'hy', 'hz']
run_from_reference.plot()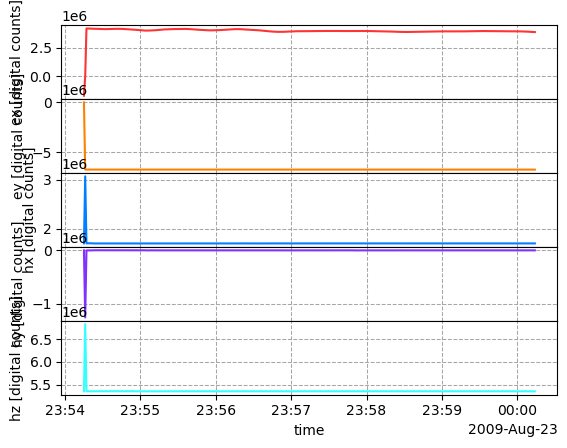

Calibrate Run¶
calibrated_run = run_from_reference.calibrate()
calibrated_run.plot()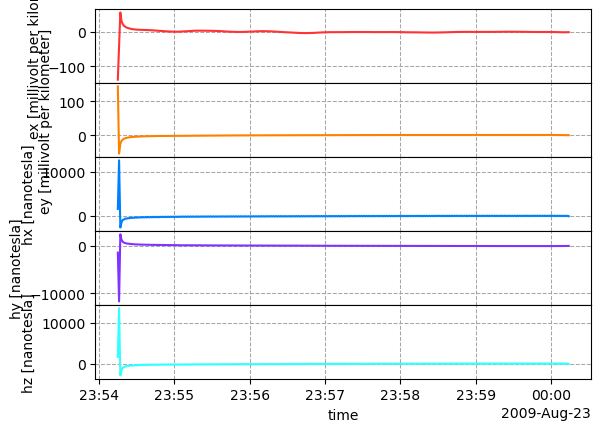

Load Transfer Functions¶
You can download the transfer functions from IRIS SPUD EMTF. This has already been done as EMTF XML format and will be loaded here.
Note that there are some discrepencies between metadata in the time series data and the transfer functions. For now we will leave it as it is, because that’s the way it is stored at IRIS, but if you really wanted to be careful you would comb through the differences and choose which one is the most correct. This is a good example of how handling metadata can be a messy job.
tf_path = default_path.parent.parent.joinpath("data","transfer_functions","earthscope")
tf_list = [tf_path.joinpath(f"USArray.{station}.2009.xml") for station in ["WYYS2", "WYYS3"]]from mt_metadata.transfer_functions.core import TFfor tf_fn in tf_list:
tf_object = TF(tf_fn)
tf_object.read()
# update run id and add new run if needed
for ii, run_id in enumerate(tf_object.station_metadata.transfer_function.runs_processed):
try:
tf_object.station_metadata.runs[ii].id = run_id
except IndexError:
new_run = deepcopy(tf_object.station_metadata.runs[0])
new_run.id = run_id
tf_object.station_metadata.add_run(new_run)
mth5_object.add_transfer_function(tf_object)2024-09-19T17:44:37.243150-0700 | WARNING | mth5.mth5 | get_survey | /Experiment/Surveys/Transportable_Array does not exist, check survey_list for existing names.
Have a look at the transfer function summary¶
mth5_object.tf_summary.summarize()
tf_df = mth5_object.tf_summary.to_dataframe()
tf_dfClose MTH5¶
We have now loaded in all the data we need for long period data. We can now process these data using Aurora and have a look at the transfer functions using MTpy.
mth5_object.close_mth5()2024-10-14T14:41:45.878984-0700 | INFO | mth5.mth5 | close_mth5 | Flushing and closing 4P_WYYS2_MTF20_WYYS3_MTC20.h5